Optuna
Optuna is a python package for tuning hyperparameters in ML. It can be integrated into such as Pytorch, Tensorflow, Keras, Scikit-learn models (more see).
In this post, I will introduce the basic workflow and give a panorama figure of Optuna. Trivial and intricate functions are remaining for interested and agog readers.
Main components
There are four components in the Optuna package:
- Hyperparameters
- Objectives
- Study
- trial
Hyperparameters
Hyperparameters are a set of parameters with different search scopes, such as the learning rate, batch size, etc. We want to find the best combination of them which achieves the optimal objective value.
Objectives
Objectives are performance criterions of potential hyperparameter combinations, such as accuracy, test loss, etc.
Trial
Trial is an instance of possible combinations where you set hyperparameters with targeted values and test and record the corresponding performance in objectives.
Study
Study is a set of trials where records all trails you have executed and the best hyperparameter combination.
Workflow
Given the set and search scopes of hyperparameters, how to choose targets values in each trial?
With a relatively small search space, enumeration may be a possible solution. However, if the parameter is continuous, how to efficiently pick values?
Fortunately, Optuna provides a Sampler class to efficiently search the space and generate possible combinations for each trial. The complicated algorithm behinds the sampling can see Optuna: efficient optimization algorithms.
Actually, if we are not researchers in this field, we don’t have to know how these sampling algorithms work, why they are efficient, etc. How to pick a suitable algorithm is more important.
In all sampling algorithms, there are two sampling types:
- Relative Sampling. In the relative sampling, there are relations among parameters and we determine values of multiple parameters simultaneously.
- Independent Sampling. In the independent sampling, parameters are independent and we determine a value of a single parameter.
The following figure illustrates the mechanism behinds Optuna.
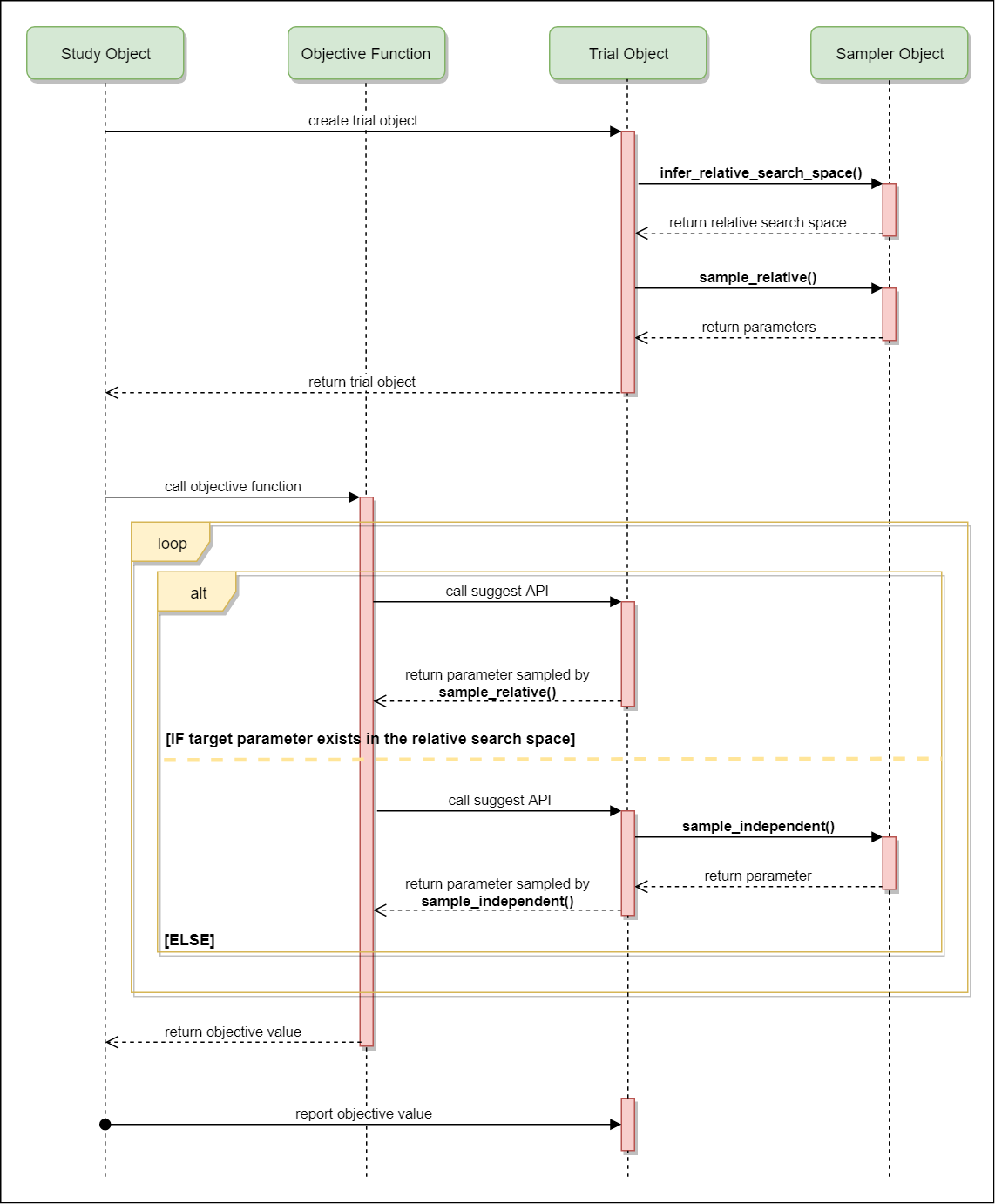
The corresponding source code is in the Trial._suggest method.
- At the beginning, A
Studyobject creates aTrialobject. When initiating theTrialobject, the constructor function invokes the pre-definedSamplerobject to determine the relative search space and carry out the relative sampling which returns parameter values of related parameters. - During test performance in a trial,
Trialprovidessuggestmethods to pick values from a parameters sequence which is generated bySimplerbased on a distribution (uniform, etc. see). - Repeat above two steps until the pre-defined trial number is achieved or error occurs. In the trial sequences, the optimization algorithm in the
Samplerwill narrow down the search space and improve efficiency.
An Example of Sampler: GridSearch
At last, I will analyze the source code of ‘GridSampler’ a derived class of Sampler.
There are three main functions in this class:
__init__sample_relativesample_independent
__init__
The most important part in the constructor is the _all_grids attribute.
def __init__(
self, search_space: Mapping[str, Sequence[GridValueType]], seed: Optional[int] = None
) -> None:
self._search_space = collections.OrderedDict()
for param_name, param_values in sorted(search_space.items()):
self._search_space[param_name] = list(param_values)
self._all_grids = list(itertools.product(*self._search_space.values()))
self._param_names = sorted(search_space.keys())
self._n_min_trials = len(self._all_grids)
self._rng = np.random.RandomState(seed)
The _all_grids attribute contains the Cartesian product of parameters in the GridSampler.
sample_relative
The sample_relative function firstly collects all unvisited grid, randomly choices one from the unvisited list and registers it as an attribute of the Study instance.
def sample_relative(
self, study: Study, trial: FrozenTrial, search_space: Dict[str, BaseDistribution]
) -> Dict[str, Any]:
target_grids = self._get_unvisited_grid_ids(study)
grid_id = int(self._rng.choice(target_grids))
study._storage.set_trial_system_attr(trial._trial_id, "search_space", self._search_space)
study._storage.set_trial_system_attr(trial._trial_id, "grid_id", grid_id)
return {}
sample_independent
The sample_independent randomly draws a grid_id from the study instance and reads the correspoding value based on the input param_name.
def sample_independent(
self,
study: Study,
trial: FrozenTrial,
param_name: str,
param_distribution: BaseDistribution,
) -> Any:
grid_id = trial.system_attrs["grid_id"]
param_value = self._all_grids[grid_id][self._param_names.index(param_name)]
return param_value